Note
Click here to download the full example code
Visualization of ROI Surface Rendered with Streamlines¶
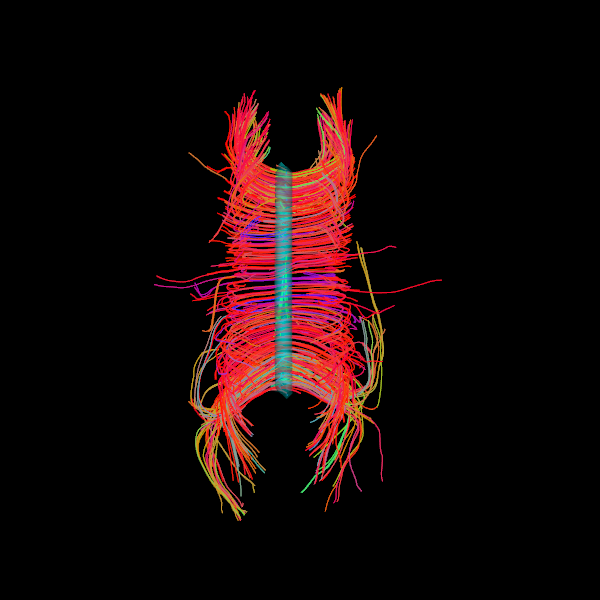
Here is a simple tutorial following the probabilistic CSA Tracking Example in which we generate a dataset of streamlines from a corpus callosum ROI, and then display them with the seed ROI rendered in 3D with 50% transparency.
from dipy.data import read_stanford_labels
from dipy.reconst.shm import CsaOdfModel
from dipy.data import default_sphere
from dipy.direction import peaks_from_model
try:
from dipy.tracking.local import ThresholdTissueClassifier as \
ThresholdStoppingCriterion
from dipy.tracking.local import LocalTracking
except ImportError:
from dipy.tracking.stopping_criterion import ThresholdStoppingCriterion
from dipy.tracking.local_tracking import LocalTracking
from dipy.tracking import utils
from dipy.tracking.streamline import Streamlines
from fury import actor, window
from fury.colormap import line_colors
First, we need to generate some streamlines. For a more complete description of these steps, please refer to the CSA Probabilistic Tracking Tutorial.
hardi_img, gtab, labels_img = read_stanford_labels()
data = hardi_img.get_data()
labels = labels_img.get_data()
affine = hardi_img.affine
white_matter = (labels == 1) | (labels == 2)
csa_model = CsaOdfModel(gtab, sh_order=6)
csa_peaks = peaks_from_model(csa_model, data, default_sphere,
relative_peak_threshold=.8,
min_separation_angle=45,
mask=white_matter)
classifier = ThresholdStoppingCriterion(csa_peaks.gfa, .25)
seed_mask = labels == 2
seeds = utils.seeds_from_mask(seed_mask, density=[1, 1, 1], affine=affine)
# Initialization of LocalTracking. The computation happens in the next step.
streamlines = LocalTracking(csa_peaks, classifier, seeds, affine,
step_size=2)
# Compute streamlines and store as a list.
streamlines = Streamlines(streamlines)
Out:
Dataset is already in place. If you want to fetch it again please first remove the folder C:\Users\skoudoro\.dipy\stanford_hardi
Dataset is already in place. If you want to fetch it again please first remove the folder C:\Users\skoudoro\.dipy\stanford_hardi
Dataset is already in place. If you want to fetch it again please first remove the folder C:\Users\skoudoro\.dipy\stanford_hardi
We will create a streamline actor from the streamlines.
streamlines_actor = actor.line(streamlines, line_colors(streamlines))
Next, we create a surface actor from the corpus callosum seed ROI. We provide the ROI data, the affine, the color in [R,G,B], and the opacity as a decimal between zero and one. Here, we set the color as blue/green with 50% opacity.
surface_opacity = 0.5
surface_color = [0, 1, 1]
seedroi_actor = actor.contour_from_roi(seed_mask, affine,
surface_color, surface_opacity)
Next, we initialize a ‘’Scene’’ object and add both actors to the rendering.
scene = window.Scene()
scene.add(streamlines_actor)
scene.add(seedroi_actor)
If you uncomment the following line, the rendering will pop up in an interactive window.
interactive = False
if interactive:
window.show(scene)
# scene.zoom(1.5)
# scene.reset_clipping_range()
window.record(scene, out_path='contour_from_roi_tutorial.png',
size=(600, 600))
Total running time of the script: ( 0 minutes 27.826 seconds)